Monte Carlo Simulations: Simulating correlated data
Independent Study / Spring 2023
Francis L. Huang, PhD
huangf@missouri.edu
@flhuang
2023.03.01 / Updated 2023-08-30
When we generate data such as this...
- We assume that the predictors are not correlated with each other (they are generated independently)
set.seed(12345); ns <- 1000x1 <- rnorm(ns); x2 <- rnorm(ns); err <- rnorm(ns)y <- 1 + .5 * x1 + .5 + x2 + errcor(x1, x2)[1] 0.03877533- This is fine but then the expected correlation of x1 and x2 = 0
- Many, times this may not be realistic
- We may want to generate correlated data (e.g, to simulate multicollinearity)
Generating correlated normal variables (from a multivariate normal distribution): bivariate case
If only two variables, can use the formula: y1=ρx1+√(1−ρ2)x2
- where ρ is the desired correlation (e.g., .5)
- x1 and x2 are two independent variables
x1 <- rnorm(10000); x2 <- rnorm(10000); rho <- .5y <- rho * x1 + (sqrt(1 - rho^2) * x2)cor(x1, y)[1] 0.5106397Generating correlated normal variables (from a multivariate normal distribution): multivariate case
# set.seed(123)# x1 <- rnorm(1000)# x2 <- rnorm(1000)# x3 <- rnorm(1000)## target correlation matrix:: what we wanttarcor <- matrix(c(1, .3, .5, .3, 1, .2, .5, .2, 1), byrow = T, ncol = 3)tarcor [,1] [,2] [,3][1,] 1.0 0.3 0.5[2,] 0.3 1.0 0.2[3,] 0.5 0.2 1.0Steps: Methods developed by Kaiser and Dickman (1962)
^Z(k×N)=F(k×k)X(k×N)
- Create a matrix of random k normal variables
- Obtain from the desired matrix a transformation matrix (F) using matrix factorization
eig <- eigen(tarcor) #has eigenvalues and eigenvectorsev <- eig$valuesevec <- eig$vectorsn <- 10000p <- 3# shortcut for just creating 3 random varsX <- matrix(rnorm(n * p), ncol = p)zhat <- t(evec %*% diag(sqrt(ev)) %*% t(X))cor(zhat) [,1] [,2] [,3][1,] 1.0000000 0.2907589 0.4978043[2,] 0.2907589 1.0000000 0.1819259[3,] 0.4978043 0.1819259 1.0000000tarcor #target correlation for comparison [,1] [,2] [,3][1,] 1.0 0.3 0.5[2,] 0.3 1.0 0.2[3,] 0.5 0.2 1.0NOTE: The original correlation matrix can be recreated using the eigenvalues and the eigenvectors
The original matrix can be decomposed into these matrices...
Σ=ΛϕΛ′ where: Λ = k × k matrix of eigenvectors and ϕ = k × k diagonal matrix with eigenvalues on the diagonal.
evec %*% diag(ev) %*% t(evec) #from previous slide [,1] [,2] [,3][1,] 1.0 0.3 0.5[2,] 0.3 1.0 0.2[3,] 0.5 0.2 1.0tarcor [,1] [,2] [,3][1,] 1.0 0.3 0.5[2,] 0.3 1.0 0.2[3,] 0.5 0.2 1.0To get the transformation matrix : Same as the principal components (signs might differ)
F=Λ√ϕ
evec %*% diag(sqrt(ev)) #transformation matrix [,1] [,2] [,3][1,] 0.8390075 0.1745109 0.5153759[2,] 0.5986054 -0.7913170 -0.1244547[3,] 0.7884457 0.4150833 -0.4539374## these are also the factor loadings from a PCA with no rotation ## where the number of components = number of variablespsych::principal(tarcor, rotate = 'none', nfactors = 3)Principal Components AnalysisCall: psych::principal(r = tarcor, nfactors = 3, rotate = "none")Standardized loadings (pattern matrix) based upon correlation matrix PC1 PC2 PC3 h2 u2 com1 0.84 -0.17 -0.52 1 -4.4e-16 1.82 0.60 0.79 0.12 1 -4.4e-16 1.93 0.79 -0.42 0.45 1 0.0e+00 2.2 PC1 PC2 PC3SS loadings 1.68 0.83 0.49Proportion Var 0.56 0.28 0.16Cumulative Var 0.56 0.84 1.00Proportion Explained 0.56 0.28 0.16Cumulative Proportion 0.56 0.84 1.00Mean item complexity = 2Test of the hypothesis that 3 components are sufficient.The root mean square of the residuals (RMSR) is 0 Fit based upon off diagonal values = 1Can do this with a variance/covariance matrix Σ:
tarcov <- matrix(c(36, -5.1, 1.8, -5.1, .96, -.34, 1.8, -.34, .25), byrow = T, ncol = 3)tarcov [,1] [,2] [,3][1,] 36.0 -5.10 1.80[2,] -5.1 0.96 -0.34[3,] 1.8 -0.34 0.25eig2 <- eigen(tarcov) #has eigenvalues and eigenvectorsev <- eig2$valuesevec <- eig2$vectorsn <- 10000p <- 3# shortcut for just creating 3 random vars; could have just # generated x1, x2, x3; variances have to be the same!X <- matrix(rnorm(n * p), ncol = p)zhat <- t(evec %*% diag(sqrt(ev)) %*% t(X))#head(zhat, n = 4)cov(zhat) [,1] [,2] [,3][1,] 36.454557 -5.1790291 1.8071540[2,] -5.179029 0.9723781 -0.3392217[3,] 1.807154 -0.3392217 0.2460346Now that you know how to do this manually, much quicker way- MASS::mvrnorm
library(MASS) # built in with R; NOTE: has a `select` function # which may conflict with dplyr::selectSimulate from a Multivariate Normal Distribution Description Produces one or more samples from the specified multivariate normal distribution.
Usage: mvrnorm(n = 1, mu, Sigma, tol = 1e-6, empirical = FALSE, EISPACK = FALSE)
Using our last example...
Need to specify the means μ as well.
tarcov [,1] [,2] [,3][1,] 36.0 -5.10 1.80[2,] -5.1 0.96 -0.34[3,] 1.8 -0.34 0.25zhat2 <- mvrnorm(n = 1000, mu = c(20, 3.2, .40), Sigma = tarcov)cov(zhat2) [,1] [,2] [,3][1,] 37.311798 -5.2984514 1.7797147[2,] -5.298451 1.0084439 -0.3467939[3,] 1.779715 -0.3467939 0.2490788- You can also specify a correlation matrix (1s on the diagonal of ∑ and covariances between -1 and 1)
- Often though, we may want to generate skewed data-- all our data so far have been normally distributed
To generate (univariate) skewed data, we can use Fleishman's (1978) Power Transformation
Y=a+bZ+cZ2+dZ3
Z∼N(0,1)
a,b,c,dare the coefficients needed to transform the unit normal variable to a non-normal variable. NOTE:a=-c
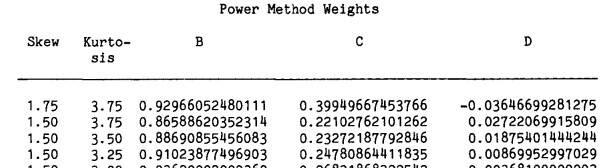
If we want a variable with a skewness of 1.75 and kurtosis of 3.75, get the coefficients from the table (p. 524)
Fleishman AI (1978). A Method for Simulating Non-normal Distributions. Psychometrika, 43, 521-532. doi: 10.1007/BF02293811
SKEWNESS KURTOSIS b c d .75 -.20 1.173302916 .207562460 -.079058576 .75 .00 1.112514484 .173629896 -.050334372 .75 .40 1.033355486 .141435163 -.018192493.75 .80 .978350485 .124833577 .001976943 .75 1.20 .935785930 .114293870 .016737509 .75 1.60 .900640275 .106782526 .028475848 .75 2.00 .870410983 .101038303 .038291124 .75 2.40 .843688891 .096435287 .046773413 .75 2.80 .819604207 .092622898 .054275030 .75 3.20 .797581770 .089386978 .06102317Example:
SKEWNESS KURTOSIS b c d1.75 3.75 .9297 .3995 -.0365set.seed(123)Z <- rnorm(1000)Y <- -.3995 + .9297 * Z + .3995 * Z^2 + -.0365 * Z^3hist(Y)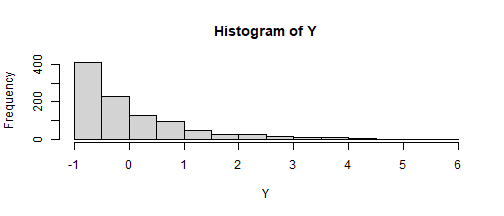
psych::skew(Y)[1] 1.765537SKEWNESS KURTOSIS b c d.75 .80 .978350485 .124833577 .001976943set.seed(123)Z <- rnorm(1000)Y <- -.12 + .98 * Z + .12 * Z^2 + .002 * Z^3hist(Y)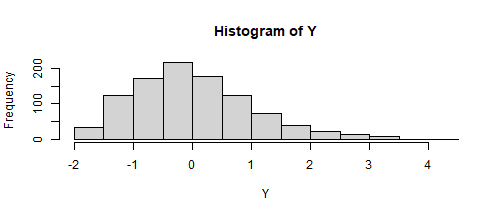
psych::skew(Y)[1] 0.7523391There is a table of coefficients in Fleishman's article: System of equations
SKEWNESS KURTOSIS b c d1.75 3.75 .9297 .3995 -.0365How do you get other coefficients?
library(SimMultiCorrData)find_constants(method = "Fleishman", skews = 1.75, skurts = 3.75)$constants c0 c1 c2 c3 -0.39949647 0.92966020 0.39949647 -0.03646691 $valid[1] "FALSE"- There are certain limits (boundaries) of the skewness and kurtosis that can be specified (cannot just specify any!). excess kurtosis ≥ skewness2 - 2 (?) (skewness-kurtosis parabola)
- O. Astivia (U Washington) has written about this (video: https://www.youtube.com/watch?v=cYx0rRBW6qA&t=4947)
- Needs a large sample to stabilize
see https://search.r-project.org/CRAN/refmans/SimMultiCorrData/html/nonnormvar1.html
find_constants(method = "Fleishman", skews = 2, skurts = 10)$constants c0 c1 c2 c3 -0.20345030 0.63044307 0.20345030 0.09874172 $valid[1] "TRUE"Z <- rnorm(10000)Y <- -.20 + .63 * Z + .20 * Z^2 + .098 * Z^3hist(Y)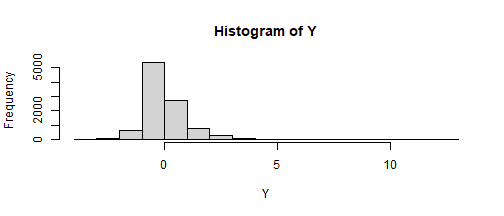
psych::skew(Y)[1] 1.947779update: Another way (not for multivariate data)-- just learned this while putting the slides together
coeff <- miceadds::fleishman_coef(mean = 0, sd = 1, skew = 2, kurt = 10)print(coeff) a b c d -0.20337308 0.63080518 0.20337308 0.09875006X1 <- miceadds::fleishman_sim(N = 10000, coef = coeff)#or... in one step...X2 <- miceadds::fleishman_sim(N = 10000, mean = 0, sd = 1, skew = 2, kurt = 10)par(mfrow = c(1, 2)) #for multipanel plot using base Rhist(X1)hist(X2)
To generate multivariate, non-normal data, cannot just use the Fleishman transformation on its own
- IMPORTANT: We can't just use the same method!
- We can generate nonnormal, independent data, then transform
- However, the correlation/covariance structure will not be exactly the same as what we expect
To generate multivariate, non-normal data, cannot just use the Fleishman transformation on its own (need Vale & Maurelli's method)
- Need to create an "intermediate" correlation matrix, then that is what is factor analyzed to get the transformation matrix, then the generated data is then transformed using the usual Fleishman coefficients (so there is an extra step). Again, we can just use a function to simplify this:
set.seed(234); par(mfrow = c(1, 1)) #resetting to 1 plotx2 <- mnonr::unonr(1000, c(1, 2), matrix(c(10, 2, 2, 5), 2, 2), skewness = c(-1, 2), kurtosis = c(3, 8))psych::pairs.panels(x2)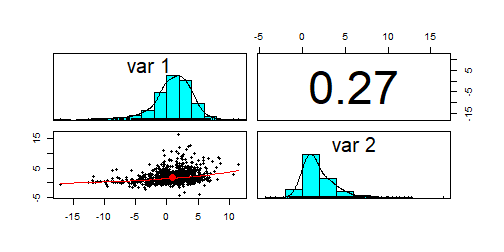
Vale, C. D. & Maurelli, V. A. (1983) Simulating multivariate non-normal distributions. Psychometrika, 48, 465-471.
Another way using the SimDesign package
library(SimDesign)set.seed(122)# univariate with skewnonnormal <- rValeMaurelli(1000, mean=10, sigma=5, skew=2, kurt=6)hist(nonnormal)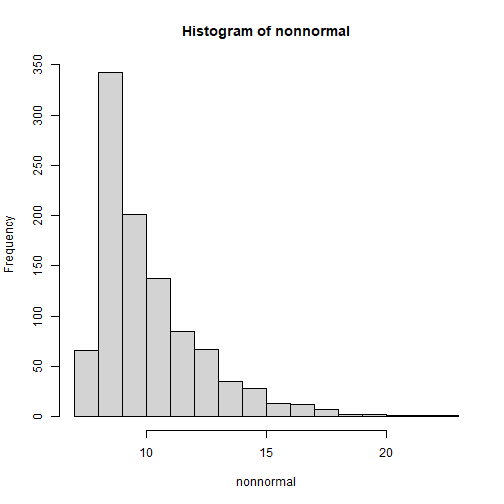
Another way using SimDesign package (cont.)
set.seed(123)n <- 10000r12 <- .4; r13 <- .9; r23 <- .1cor <- matrix(c(1,r12,r13, r12,1,r23, r13,r23,1),3,3)sk <- c(1.5,-1.5,0.5)ku <- c(3.75,3.5,0.5)nonnormal <- rValeMaurelli(n, sigma = cor, skew = sk, kurt = ku)hist(nonnormal[,1])
hist(nonnormal[,2])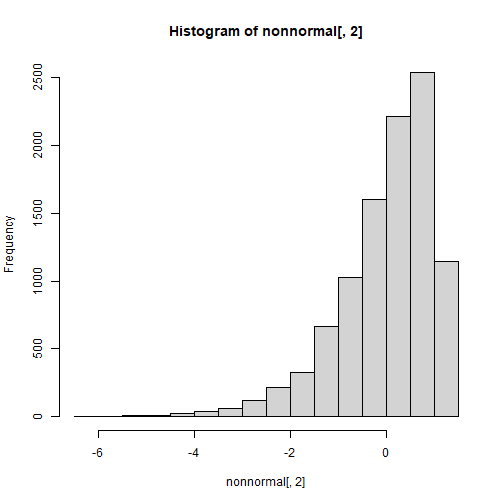
hist(nonnormal[,3])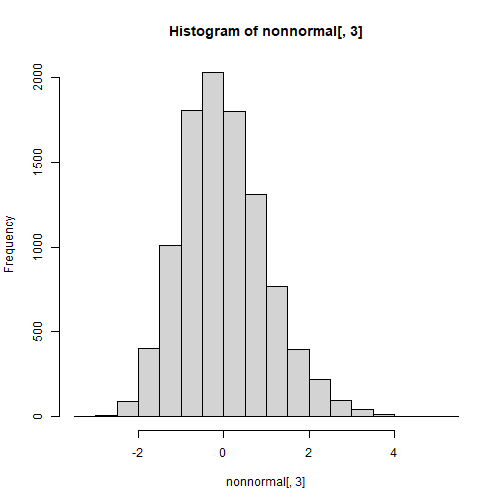
Sidenote: At times, the covariance is specified
... so need to just convert and the correlation is .28. This is just: R=S−1ΣS−1
mm <- matrix(c(10, 2, 2, 5), ncol = 2)(sdm <- diag(sqrt(diag(mm)))) #diagonal matrix with sd on the diagonal [,1] [,2][1,] 3.162278 0.000000[2,] 0.000000 2.236068Sinv <- solve(sdm)Sinv %*% mm %*% Sinv #correlation matrix [,1] [,2][1,] 1.0000000 0.2828427[2,] 0.2828427 1.0000000Or can simply use:
cov2cor(mm) [,1] [,2][1,] 1.0000000 0.2828427[2,] 0.2828427 1.0000000There are several uses of this (generating multivariate correlated data)
- You can run regressions using a correlation/covariance matrix
- You can simulated CFA/SEM models too (w/c we haven't done)
e.g., a common factor model
Can be shown as:
Σ=ΛΦΛ′+Θ
where:
- Σ = population correlation matrix (Sigma)
- Λ = population factor loading matrix (Lambda)
- Φ = population factor correlation matrix (Phi)
- Θ = unique variances matrix (Theta)
Simulating a two factor model: Setting up the matrices...
Two factor model:
- The standardized loadings are .6 using two items each
- No cross loadings
- F1 and F2 are correlated at .5
Lam <- matrix(c(.6, 0, .6, 0, 0, .6, 0, .6), ncol = 2, byrow = T)Phi <- matrix(c(1, .5, .5, 1), byrow = T, ncol = 2)Theta <- diag(.64, 4)cr <- Lam %*% Phi %*% t(Lam) + Thetacolnames(cr) <- rownames(cr) <- paste0("X", 1:4)cr X1 X2 X3 X4X1 1.00 0.36 0.18 0.18X2 0.36 1.00 0.18 0.18X3 0.18 0.18 1.00 0.36X4 0.18 0.18 0.36 1.00Estimate using lavaan... recovers what we specified (since we are working from the matrices)
library(lavaan)mod <- ' f1 =~ X1 + X2 f2 =~ X3 + X4'm1 <- cfa(model = mod, sample.cov = cr, sample.nobs = 1000 )summary(m1, standardized = T)Latent Variables: Estimate Std.Err z-value P(>|z|) Std.lv Std.all f1 =~ X1 1.000 0.600 0.600 X2 1.000 0.164 6.101 0.000 0.600 0.600 f2 =~ X3 1.000 0.600 0.600 X4 1.000 0.164 6.101 0.000 0.600 0.600Covariances: Estimate Std.Err z-value P(>|z|) Std.lv Std.all f1 ~~ f2 0.180 0.030 5.902 0.000 0.500 0.500Variances: Estimate Std.Err z-value P(>|z|) Std.lv Std.all .X1 0.639 0.065 9.818 0.000 0.639 0.640 .X2 0.639 0.065 9.818 0.000 0.639 0.640 .X3 0.639 0.065 9.818 0.000 0.639 0.640 .X4 0.639 0.065 9.818 0.000 0.639 0.640Latent Variables: Estimate Std.Err z-value P(>|z|) Std.lv Std.all f1 =~ X1 1.000 0.600 0.600 X2 1.000 0.164 6.101 0.000 0.600 0.600 f2 =~ X3 1.000 0.600 0.600 X4 1.000 0.164 6.101 0.000 0.600 0.600Covariances: Estimate Std.Err z-value P(>|z|) Std.lv Std.all f1 ~~ f2 0.180 0.030 5.902 0.000 0.500 0.500Variances: Estimate Std.Err z-value P(>|z|) Std.lv Std.all .X1 0.639 0.065 9.818 0.000 0.639 0.640 .X2 0.639 0.065 9.818 0.000 0.639 0.640 .X3 0.639 0.065 9.818 0.000 0.639 0.640 .X4 0.639 0.065 9.818 0.000 0.639 0.640Can generate simulated data then using the matrices (using the methods we discussed)...
- Then you can evaluate the performance of your model
- Examine fit indices?
- Results of misspecification?
- How does it deal with skewed data?
- What about the use of different estimators?
- What about ordinal data? (we haven't discussed that)
Others...
Another 'newish' package for generating simulated data is the faux (deBruine) package
- It looks interesting and has nice documentation

Others: generating categorical data with lavaan
library(lavaan)# setup population model (from Sonja W)# the x1 | -.6*t1 + .6*t2 specifies that x1 has two thresholds # (so three categories) set at -.6 and .6. lavaan uses delta # parameterization by default, so these thresholds# cut up a standard normal distribution in 3 sections that map on to# the observed categorical responsespopmod <- 'f1 =~ .8*x1 + .8*x2 + .8*x3 + .8*x4x1 | -.6*t1 + .6*t2 + 1*t3x2 | -.6*t1 + .6*t2 + 1*t3x3 | -.6*t1 + .6*t2 + 1*t3x4 | -.6*t1 + 0*t2 + .6*t3'simdat <- simulateData(model = popmod, model.type = "cfa", std.lv = T, sample.nobs = 200)added: 2023.08.30
psych::headTail(simdat) x1 x2 x3 x41 2 2 2 22 1 2 2 33 3 2 2 34 4 2 4 4... ... ... ... ...197 1 2 1 3198 3 2 2 4199 2 2 2 3200 2 2 1 3table(simdat$x1) 1 2 3 4 70 80 19 31table(simdat$x4) 1 2 3 4 61 49 47 43